基因组的可塑性增强了屎肠球菌的环境适应性
时间:2019-08-22 来源:益生之源 作者:益生之源 浏览次数:2531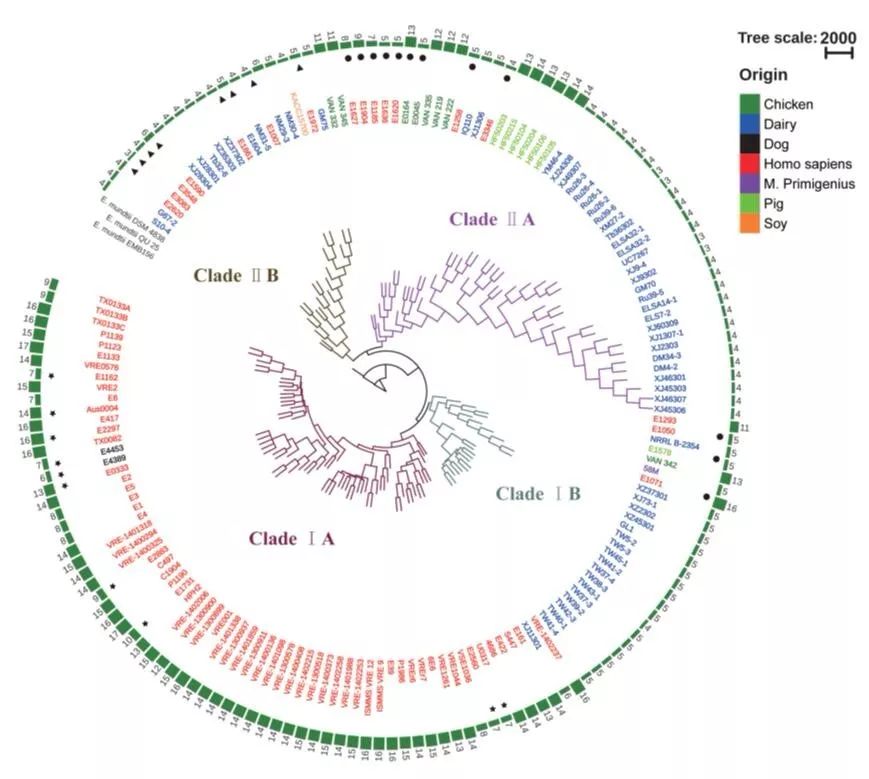
Figure 4:Phylogenetic tree constructed based on the core genes of Enterococcus (E.)faeciumisolates. The phylogenetic tree wasconstructed using the DNA sequences of 871 core genes of 161 isolates ofE. faecium and 3 isolates of E. mundti. Enterococcus mundtii is the closest phylogenetic relative of E. faecium and was thus included asoutgroup. The color of the isolate name represents the origin of theisolate. Isolates labeled with star, dot, and triangle in the out circle wereclustered in clade A1 (hospital-associated), clade A2 (animal-associated), andclade B (commensal-associated), respectively, in Lebreton et al [9]. The numberof antibiotic resistance genes was labeled in the out circle.
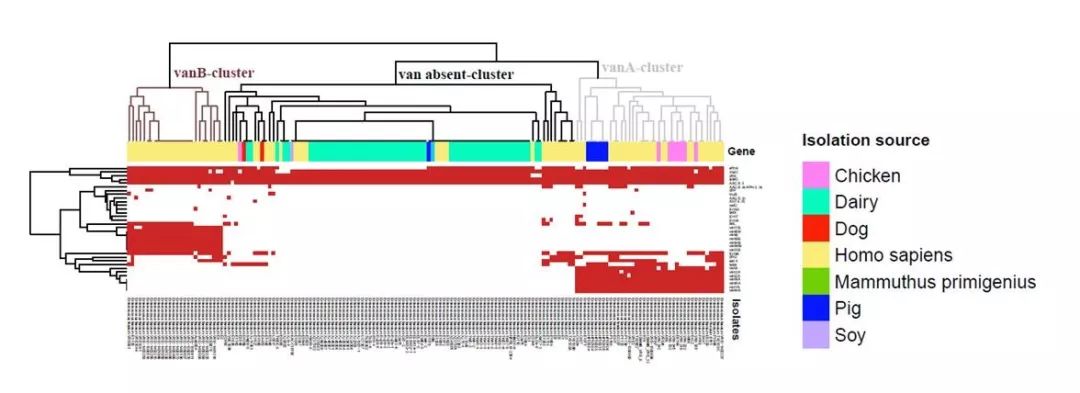
Figure 5: Clusteranalysis based on the profile of antibiotic resistance genes.
Red dots represent genes present in the individualisolates.Threeclusters could be identified based on the distribution of vancomycin resistancegenes: vanA-cluster, vanB-cluster, and van absent-cluster.
免责声明:本文仅代表作者个人观点,与中国益生菌网无关。其原创性以及文中陈述文字和内容未经本站证实,对本文以及其中全部或者部分内容、文字的真实性、完整性、及时性本站不作任何保证或承诺,请读者仅作参考,并请自行核实相关内容。
版权声明
1.本站部分转载的文章非原创,其版权和文责属于原作者。2.本网所有转载文章、链接及图片系出于传递更多信息之目的,且明确注明来源和作者,不希望被转载的媒体或个人可与我们联系,对可以提供充分证据的侵权信息,bio149将在确认后12小时内删除。3.欢迎用户投递原创文章至86371366@qq.com,经审核后发布到首页,其版权和文责属于投递者。







